Task Description
The images in the proposed dataset are all at the line level and annotated with Chemfig strings. This competition comprises a single task aimed at transcribing the correct Chemfig string. Given the non-uniqueness in Chemfig representations, we will employ graph matching algorithms to assess whether the predicted and target Chemfig strings match.
Evaluation Metrics
Exact Match (EM)
Similar to handwritten mathematical expression recognition tasks, we use the EM score as the primary evaluation criterion. Specifically, EM indicates a complete match between the predicted and labeled strings. For the Chemfig modeling approach, given the polysemous nature of the labels, we first convert both the labels and recognition strings into graphs and then determine if their graph forms match with a Graph Matching Tool. Let T denote the number of samples and R denote the number of predicted results that match the labeled results, then EM can be calculated using the following formula:
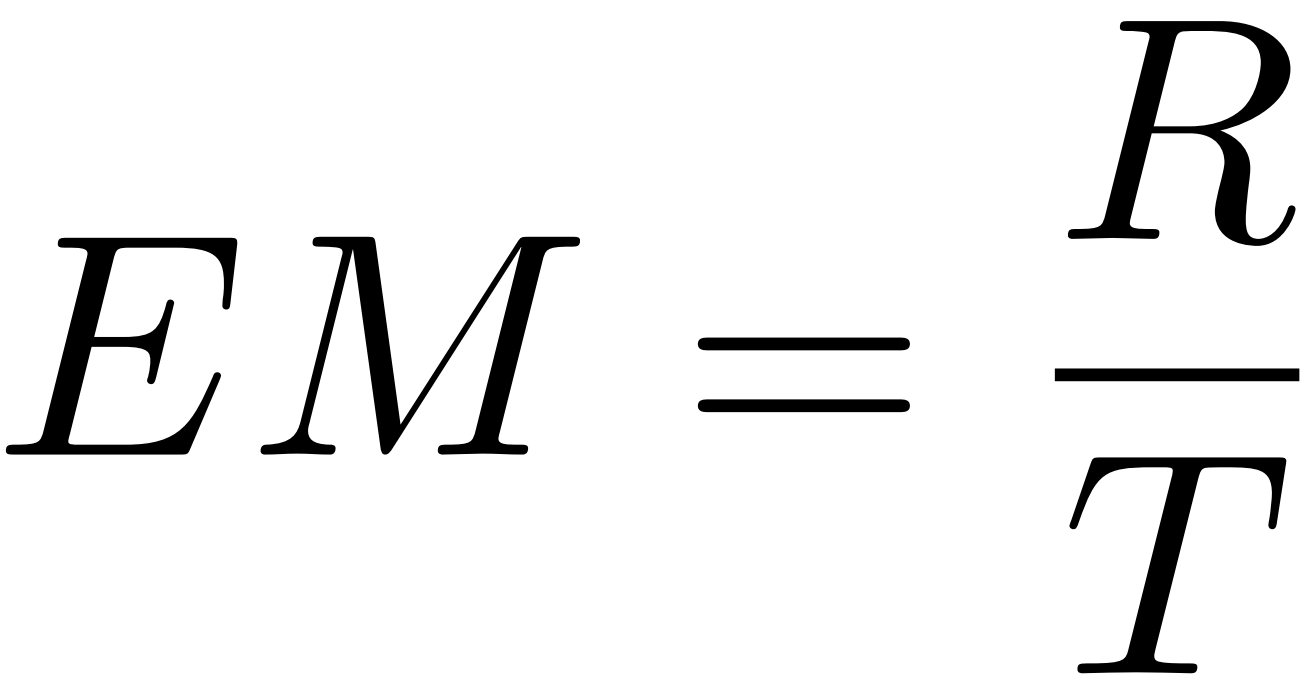
In addition, for our EDU-CHEMC data, since it contains mixtures of formulas and molecular structures, a single image may contain multiple molecular structures. Therefore, we define another auxiliary metric as follows.
Structure Exact Match (Structure EM)
For samples that contain mixed molecular structure and regular formulas, when all the molecular structure recognition results match the labeled graphs, we consider that the molecular structure of the sample has been “correctly recognized”. Let T denote the number of samples and Rstruct denote the number of samples with correctly recognized structures, then:
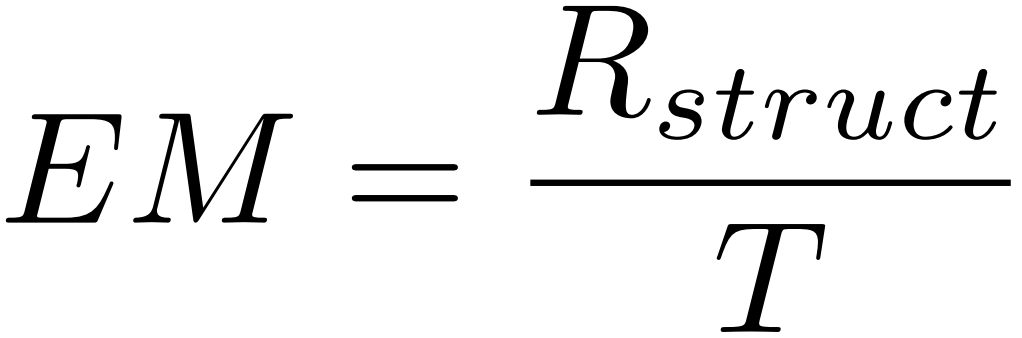
Structure EM measures the model’s recognition performance for the molecular structures in the mixed-mode data exclusively.
In the ranking protocol of this competition, we initially compare the EM (Exact Match) metric. In cases where two teams have the same EM scores, the final ranking will be determined based on their respective Structure Exact Match (Structure EM) scores.
Graph Matching Tool
Here, we offer a brief introduction of the Graph Matching Tool.
It firstly extract all chemfig regions of a recognition result or a label string. For tokens outside chemfig regions, it directly compare the result string and label string, and only exact match will be considered to be correct. For tokens inside a chemfig region, it parse the content inside this chemfig region to graph, and then use a heuristic method to judge whether the graphs of label and recognition result are matched.
The rules for graph matching are as follows:
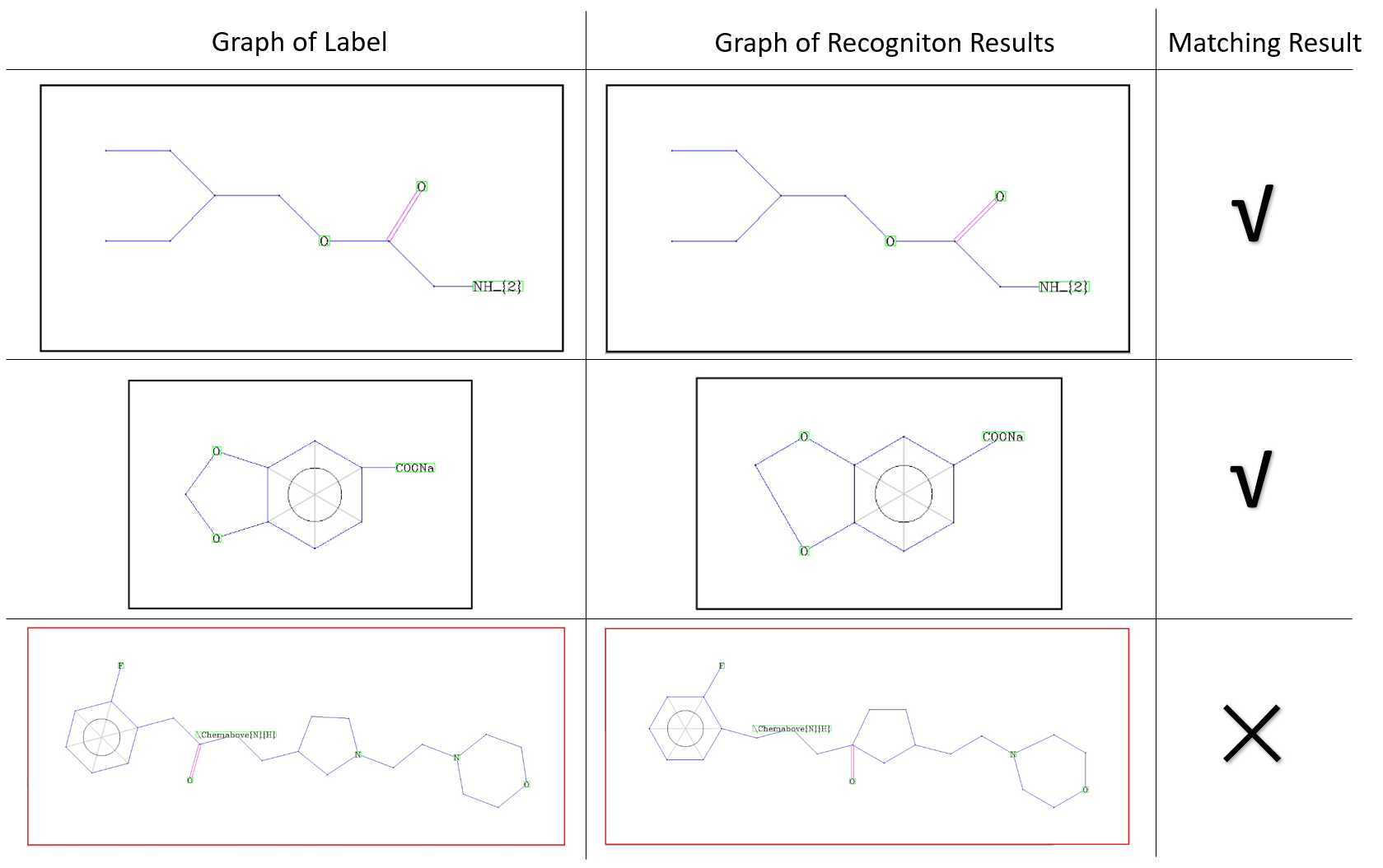
- All atoms must be correctly recognized, meaning the tokens of all atoms should match the label.
- Types of all bonds should be correctly recognized.
- Bond angles do not need to be an exact match, but the deviation should not be too large. Angles are used to recover the structure and appearance from string tokens.
- All connection relationship between atoms must be correctly recognized.
- Overall, the structure and appearance of the recognition result's graph should match the label.
Refer to Figure 1 for examples of graph matching results.